TCRISPR is a simple acronym to remember, but it refers to a very complex technology that promises to revolutionize gene editing and splicing. The letters stand for “clustered regularly interspaced short palindromic repeats”. The palindromic repeats are short repeated DNA sequences found in some bacteria that geneticists have been aware of since 1987, but it took 20 years to realise they are part of the bacteria’s defense system against viruses. In 2013 scientists achieved some key breakthroughs, and are now able to mimic and modify the naturally occurring CRISPR bacterial immune mechanism, and harness it for specific purposes.
Overview
In crude terms, this is how the CRISPR defense mechanism works in bacteria:
- Within the bacterial cells are enzymes capable of chopping up DNA, and these are called CRISPR associated proteins (‘Cas’).
- When a bacterium is invaded by a virus, or more correctly a bacteriophage, Cas snip the viral DNA into short sections, and then store those sections in the CRISPR region of their own DNA.
- Over time, many of these copies of viral DNA bits are stored, and are separated by repeated spacers (the palindromic repeats).
- Each viral DNA sequence is then copied into an RNA molecule.
- A Cas then captures each RNA molecule, and floats around within the cell.
- When a Cas encounters viral genetic material that matches the DNA sequence it is carrying, it latches on to it.
- Once attached to the viral DNA, the Cas chops it in half, thus rendering the viral DNA incapable of replication.
Simplistically, each Cas is given a genetic description of the “villain” it should target, then roams around until it identifies and incapacitates its particular designated “villain”. This fundamental mechanism has now been harnessed for human purposes.
Detailed Description
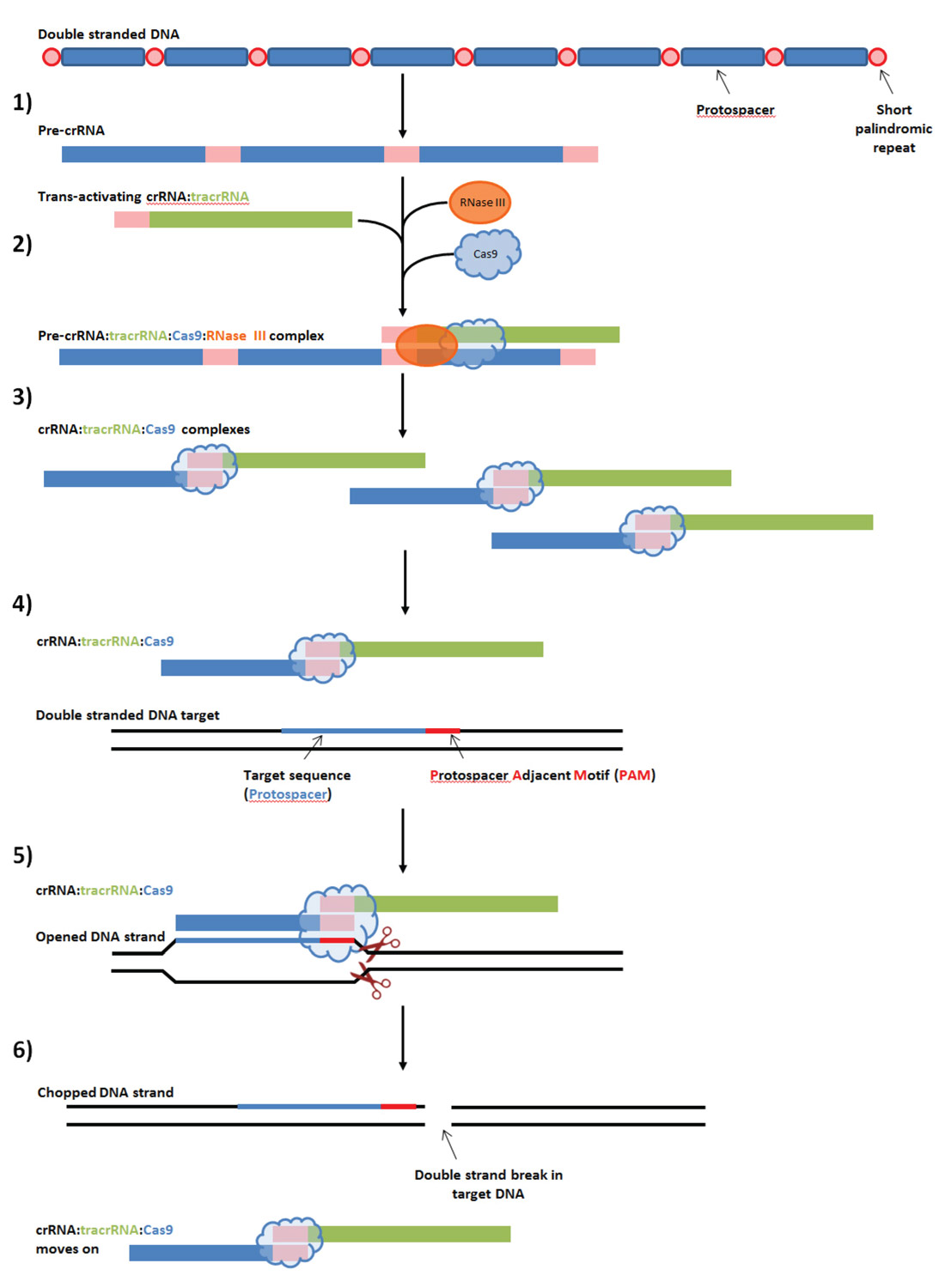
Figure 1 shows the main steps involved in the naturally occurring CRISPR immune system.
- The entire CRISPR array made up of many protospacers and short palindromic repeats is transcribed, to form the pre-crRNA sequence.
- Special trans-activating crRNAs (tracrRNA) process the pre-crRNA into separate crRNAs. The tracrRNA possesses a protein sequence similar (homologous) to the short palindromic repeats, and is thus able to identify where each protospacer starts and ends; it then recruits RNase III and Cas9 enzymes which do the actual separation.
- Once the separation is done, a multitude of crRNA:tracrRNA:RNase III complexes are ready for action.
- The complexes float around, seeking viral DNA sequences that match their crRNA. Note that with the Type II Cas9 system that science is modifying and using, a target is only valid if a unique protospacer adjacent motif (PAM) is present immediately after the target DNA.
- Once a target virus DNA is identified, the Cas binds to it using the PAM to position itself correctly, separates the two DNA strands, and chops them both apart right after the PAM.
- After the chopping is done, the crRNA:tracrRNA:RNase III complex unbinds, and the viral DNA is left incapable of replication.
Applications
Prior to 2013, there were two technologies available for gene editing: transcription-activator-like effector nucleases (TALEN) and zinc finger nucleases (ZFN). Both are relatively clumsy and time consuming because they require custom designed protein nucleases that bind to specific targeted genomes. Once CRISPRs were identified as capable of modifying DNA in a targeted fashion, researchers sought to harness them. There are three Types of bacterial CRISPRs, I, II, and III; Type II is the one that scientists have mainly had success with.
The CRISPR system being used is relatively easy (compared to TALEN and ZFN) to generate and deploy. Like the naturally occurring bacterial version it mimics, it is made up of two parts: a guide RNA (gRNA) and a non-specific CRISPR-associated endonuclease known as Cas9. There have been about 45 Cas identified, and Cas9 happens to be the one that is most amenable to modification. However, CRISPRs with slightly different mechanisms and attributes are being identified and used – for example, the Cpf1 is smaller, identifies a different PAM, and its cuts are slightly offset; there is some hope it can be used to knock out malaria DNA. The gRNA consists of a scaffolding structure of sorts into which a short (~20 nucleotides) target DNA protospacer sequence is inserted. Both the scaffolding part of the gRNA and the Cas9 endonuclease are easy to produce, so all that is required to use the system is to insert the desired target DNA sequence into the scaffolding. Companies are producing all the required components, including many different target protospacers, and offering them commercially. The website I used as my main resource (www.addgene.org) is a good example of the commercial state of the business.
Scientists first used the CRISPR-Cas9 system to knock out specific parts of a DNA sequence. The medical research community was quick to see the potential and a huge effort quickly resulted in modifications that allowed several new functionalities that are revolutionizing the field of genetic modification. These include CRISPRs that can screen, activate, repress, image and purify gene sequences.
DNA possesses enzymes capable of repairing damage to the DNA, and these are exploited when DNA segments are removed and replaced – the unwanted bits are snipped out and replaced with a new sequence, then the DNA heals. The offset cuts produced by the previously mentioned Cpf1 tend to heal better, and this is the sort of advantage scientists are aggressively pursuing. To achieve other functionalities, scientists did something counterintuitive – they disabled the Cas’ snipping ability. This made the Cas available to carry molecules designed to perform other tasks – repress, activate, purify and image. I find the latter intriguing; here the “dead” Cas carries phosphorescent molecules that illuminate the targeted DNA, perfect for certain kinds of research.
My original intention was to delve more deeply into each of the above-mentioned six applications, but I quickly realized that the science involved is well beyond my crude understanding of genetic chemistry, or chemistry period. Suffice to say, in all of the applications the CRISPR binds to the target, and then the action of the Cas is dependent on its desired functionality. I invite interested readers to pursue the topic on their own and have provided some references, both light and heavy, to facilitate this.
Implications
CRISPR represents a huge double edged genetic sword. On the one hand there exists the possibility that CRISPR technology could lead to the eradication of diseases and genetic defects that have plagued humans (and other creatures) since the beginning of time. On the other hand the technology could be used to do all sorts of unethical things, such as unleash horrific bio-medical weapons designed to modify people’s genetic material irreparably; it could also be used to perform ethically questionable things like modify a baby’s genetic characteristics while still in the womb. We think performance enhancing drugs in sport are bad, but imagine if perfect NFL specimens and the like were being created at the fetal stage! And instead of plastic surgery, people could be modified to be physically perfect before birth. What if genes associated with aging are identified and understood? I could have a magnificent mane of hair like Kim Jong-un and live forever! Bwah-hah-hah! I predict that a James Bond villain in possession of CRISPR technology is just around the corner.
The number of possible applications – good, bad and questionable – is almost limitless. It begs the question whether in addition to the scientific work being done, study and careful thought should not be put into preparing and arming our legal system to ethically govern the use of these emerging genetic technologies.










Share This Column